1. Introduction¶
1.1. What is ExTASY ?¶
ExTASY, the Extensible Toolkit for Advanced Sampling and analYsis, is a flexible toolkit to allow efficient sampling of complex macromolecules using molecular Dynamics in combination with on-the-fly analysis tools, to drive the sampling process to regions of interest. In particular, compared with existing approaches like metadynamics, ExTASY requires no a priori assumptions about the behaviour of the system. ExTASY consists of several interoperable Python tools, which are coupled together into pre-defined patterns that may be executed on compute resources ranging from PCs and small clusters, to large-scale HPC systems.
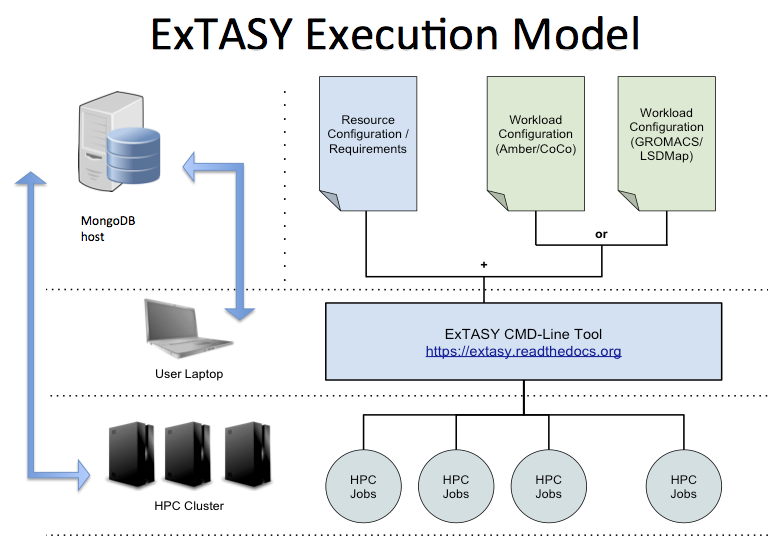
ExTASY provides a command line interface, that along with specific configuration files, keeps the user’s job minimal and free of the underlying execution methods and data management that is resource specific. The ExTASY user interface is run on your local machine and handles the data staging, job scheduling and execution on the target machine in a uniform manner, making it easy to test small systems locally before moving to larger HPC resources as needed.
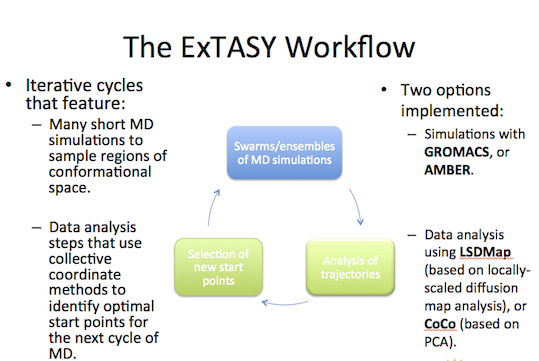
The coupled simulation-analysis execution pattern (aka ExTASY pattern) currently supports two usecases:
- Gromacs as the “Simulator” and LSDMap as the “Analyzer”
- AMBER as the “Simulator” and CoCo as the “Analyzer”
1.2. The ExTASY approach¶
ExTASY uses swarm/ensemble simulation strategies that map efficiently onto HPC services. It uses smart collective coordinate strategies to focus sampling in interesting regions, and relies on machine learning methods rather than user expertise to select and refine (on the fly) the collective coordinates. ExTASY is compatible with standard MD codes out of the box - without requiring software patches.
1.3. Background¶
1.3.1. Why do enhanced sampling ?¶
To efficiently and accurately identify particular alternative conformations of a molecule.
- E.g., starting from an apo-conformation, identify alternative low-energy conformations of a protein relevant to ligand binding (induced fit/conformational selection).
To efficiently and accurately sample ALL conformational space for a molecule.
- E.g., calculation of thermodynamic and kinetic parameters.
1.3.2. How to do enhanced sampling ?¶
Faster MD through hardware and software developments, e.g.:
- multicore architectures and domain composition.
- specialized hardware (ANTON, GRAPE,...).
Faster MD through manipulation of the effective potential energy surface, e.g.:
- meta-dynamics,
- accelerated dynamics.
Faster sampling through multiple simulation strategies, e.g.:
- Replica exchange.
- Swarm/ensemble simulations and Markov chain models.